Oncogenomics
Research activities
The laboratory of Oncogenomics (ISPRO-CRL) is located in Pisa, at the Institute of Clinical Physiology of CNR, and is focused on the study of melanoma, pursuing the following lines of research.
1. Study of BRAFV600E isoforms
We have recently discovered that, contrary to what reported in databases, BRAFV600E does not exist as a single protein and a single mRNA encoding for it. It rather exists as a pool of protein and mRNA isoforms that differ in length and sequence. Currently, we are using multiple experimental approaches in order to understand if and how these differences contribute to the post-transcriptional regulation of BRAFV600E expression and to the functionality of BRAFV600E protein. These findings carry important therapeutic implications, as they might guide the development of more specific hence more effective BRAF inhibitors.
2. Identification and study of new functional interactors of BRAFV600E
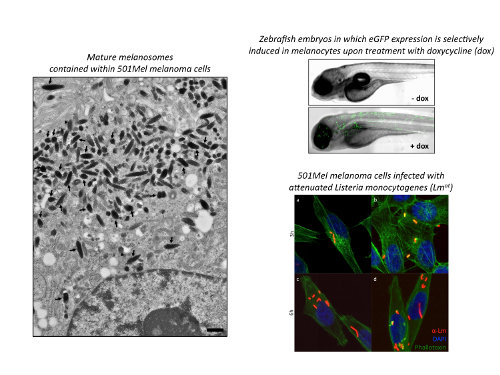
Once established that in yeast S. cerevisiae human BRAF can exert its kinase activity, we decided to use this model system in order to identify, through genome-wide screening, new functional interactors of BRAFV600E, i.e. proteins that, when absent or overexpressed, can alter its functionality. The oncosuppressive/oncogenic features of such proteins are confirmed in vitro (melanoma cell lines) and in zebrafish, using transgenic strains that develop melanoma. Next, we study the relationship between functional interactors and BRAFV600E at the molecular level. Finally, we assess the relevance of BRAFV600E functional interactors for human melanoma. With these studies, we aim to discover proteins that, if targeted together with BRAFV600E, can potentiate the efficacy of BRAF inhibitors.
3. Study of BRAFV600E-regulated microRNAs
By means of small RNA sequencing, we have identified a group of microRNAs whose levels vary in melanoma cells upon treatment with vemurafenib, an inhibitor of BRAFV600E kinase activity. The functional analysis of a subset of these microRNAs was performed in vitro (melanoma cell lines) and in vivo, using xenograft in zebrafish as well as in immunocompromised mice. In this way, we were able to establish that they affect multiple aspects of melanoma cell biology, including motility and pigmentation, and, in so doing, they favor or inhibit vemurafenib activity. We are currently generating an inducible melanoma model in zebrafish, so that we can further study, in vivo, the role played by our microRNAs of interest in melanomagenesis. We are also setting up a preclinical model in immunocompromised mice, in order to test whether the modulation of the functions of our microRNAs can potentiate the efficacy of BRAF inhibitors.
4. Development of a new strategy for the selective delivery of drugs inside melanoma cells
New targeted anticancer drugs are constantly developed, but side effects (not only cancer cells but also normal cells are targeted) remain one of their main drawbacks. To overcome this problem, we are developing an innovative strategy for drug delivery. This strategy is based on an intracellular bacterium that, in its attenuated form, shows selective tropism for cancer cells, both those located at primary tumor sites and, critically, those located at metastatic sites. Furthermore, this bacterium can be adapted to carry single drugs as well as drug combinations inside melanoma cells. Currently, we are testing our delivery strategy both in vitro (melanoma cell lines) and in vivo, using a genetically engineered mouse model in which melanoma tumors develop in an inducible and tissue-specific fashion.














